A successful surveillance program for antibiotic resistant Escherichia coli in Australia should account for the heterogenous nature of the food-animal population. Studies that rely on measurements made on several hundred isolates can only satisfy limited objectives because they risk imprecise and biased estimation of the presence and distribution of resistance traits. Observations on a larger number of isolates are needed to ensure animal, herd and region effects are adequately represented so that findings can be extrapolated to the appropriate population of interest. An efficient methodology for measuring the resistance traits of a large number of isolates is described. Commun Dis Intell 2003;27 Suppl:S117-S120.
Top of page
Introduction
Generic indicator organisms such as Escherichia coli play a prominent role in many existing surveillance systems for antibiotic resistance in animals.1E. coli is regarded as a useful indicator of antibiotic resistance in the bacterial flora of livestock and food because it responds to the selective pressures of antibiotics, because it is ubiquitous in the gut of food animals, and because it readily persists in raw foods and the environment. Individual E. coli isolates are easily studied in the laboratory to yield results that can be interpreted using standard criteria. E. coli is therefore a strong candidate for inclusion in future studies of the spatial and temporal distribution of antibiotic resistance in Australian livestock and livestock products.
Practical considerations
Future surveillance systems for antibiotic resistance will need to allow for the extreme heterogeneity that characterises livestock production in Australia. There are a plethora of animal species, animal breeds and management systems located within many different climatic regions. Hence, food-animal production occurs in diverse environments resulting in variable degrees of exposure to possible sources of resistant organisms (other herds or flocks, humans, wildlife and environmental contamination). Even at a particular locality and within a specific industry there can be large differences in husbandry practices. Antibiotic usage patterns range from continuous inclusion of low-concentrations in rations in some intensive production systems to extremely rare, highly selective or even absent in the extensive beef and sheep grazing industries.
The underlying complexity of the animal industries impacts on the ways in which surveillance for resistant E. coli could be conducted. In particular, the low profitability of some production systems and the low monetary value of most individual animals explains why veterinary laboratories receive only a small number of requests for antibiotic resistance testing of E. coli isolates from food animals. Moreover, the selective nature of veterinary diagnostic submissions means that the E. coli isolates obtained from diagnostic testing are not likely to be representative of those entering the food chain. Thus, passively acquired data appear to be poorly suited to accommodating the complexity present in the underlying population of E. coli derived from animals. Purposefully designed surveys of the livestock population and livestock products are best suited to providing data forming a basis for national policies.
Top of page
Implementation of active surveillance
Active surveillance for resistant E. coli in animal populations should be designed and analysed to account for the sources of variation in the population of isolates and valid confidence limits for the proportion of E. coli with a specific resistance trait can be generated. Confidence limits need only be as narrow as required by the study objectives which in turn should reflect how the surveillance findings will contribute to decision making. It is also a requirement that the strength of association between risk factors and the occurrence of resistance be estimated without bias. To meet these aims a number of sampling issues need to be addressed. One of these is the requirement to test a sufficient number of isolates, a sufficient number of animals, herds (flocks), and at a sufficient number of points in time to allow firm inferences to be made about the distribution of resistance and sources of variation. A second requirement is the need to ensure that sampling is performed to account for the likely 'contagious' or 'clustered' pattern of distribution of resistant isolates. A third consideration is to design sampling methods that allow confidence interval estimates of prevalence to be produced that are based on all of the sources of variation in the population. The latter requirement can usually be satisfied by using statistical techniques for estimation of variance components provided an appropriate study design is implemented.2 However, the first two of these requirements have hitherto been difficult to satisfy because the high cost of assessing a large number of E. coli isolates has restricted the options available for study design. Thus, only small numbers of herds, product consignments, or regions appear to be represented in most existing data and it is difficult to extrapolate the findings beyond those animals or products included in the study.
To illustrate the importance of sample size and sampling error one can make the simplifying assumption that the resistance phenotypes of interest occur at random throughout the populations of E. coli, animals and herds. The required sample size for estimating prevalence can then be calculated from the binomial probability distribution. If the objective is to estimate the prevalence of tetracycline resistance (at a particular concentration) amongst E. coli isolates from intensively-raised animals, such that the 95 per cent confidence limits are each within 5 per cent of the point estimate, then about 400 isolates would require evaluation (this assumes the expected prevalence is close to 50 per cent -an assumption that is justified on the basis of published estimates from pigs and poultry,3,4,5 and because it is appropriate to err closer to 50 per cent to ensure a sufficient sample size). The required sample size is between 1.3 and 2 times greater that number relied upon in various national surveys to assess the proportion of resistant E. coli from a single animal species or product.4,5,6 Moreover, this sample size calculation is probably an underestimate because it assumes that the only variation is due to random error, that is, it takes no account of the likely non-random distribution of resistance isolates in the population of interest.
'Clustering' (likeness amongst observations close in time or space) is a term used to describe the non-random source of variation that is commonly associated with the distribution of infectious agents or disease events. In the case of antibiotic resistance in animal populations, clustering could feasibly be induced by a range of determinants of resistance (known or unknown) such as exposure to antibiotics in rations or exposure to human effluent contaminating the environment. In veterinary epidemiology clustering is often exaggerated because of the way that animals are managed in commercial units (herds or flocks), and by the way that herds and flocks at a similar geographic location tend to have a common set of risk factors.7 Sometimes clustering occurs for no obvious reason and this has been shown to be the case for the clustering of particular resistant E. coli phenotypes within individual pigs housed in the same pen.8 The pervasiveness of clustering prevents the indiscriminate use of binomial probabilities (or any approximations thereof) for generating confidence limits or sample sizes. Unfortunately, the estimation of sample sizes (number of isolates, number of animals, number of herds or flocks etc.) is difficult in the absence of any estimates of variance. The number of isolates required to be evaluated could be several times greater than what is predicted by the approach described above. Consequently, during the initial phases of surveillance there is a need to estimate components of variance attributable to the different levels of sampling (isolates, animals, herds, or flocks etc.) to aid in the design of subsequent sampling plans.
Top of page
New methodology
To overcome the difficulties of study design and sampling, a laboratory technique for assessing large numbers of E. coli isolates per specimen has been adopted in a pilot study of dairy cattle in north-eastern New South Wales. The aims are to develop methodology that could be used to efficiently study the distribution of resistance in animals, humans and environmental samples. The approach has been adapted from work on E. coli in pig populations in Canada3 and is based on hydrophobic grid membrane filtration (HGMF). Key elements of the procedure are: the growth of E. coli by filtering diluted animal faeces through HGMF grids, incubation of grids on selective agar, replication of colonies onto HGMF grids that are either incubated on chromogenic agar (for E. coli identification) or on agar containing antibiotics and made to standard specifications.9 The final and critical step for achieving economy of scale is to use computer software to perform image analysis of HGMF grids to detect the growth of E. coli, to compute multiple resistance patterns for each isolate, and to incorporate the findings for each specimen in a surveillance database. The process is summarised in the Table.
Table. Summary of steps performed during antibiotic resistance testing of Escherichia coli derived from cattle faeces using the HGMF procedure and image analysis
| Step 1. |
Fresh cattle faeces obtained during farm visits. |
| Step 2. |
Tenfold serial dilution of specimens prepared and stored. |
| Step 3. |
Preliminary estimation of the concentration of E. coli per
gram of faeces determined by spread plate or HGMF enumeration technique. |
| Step 4. |
A master HGMF grid is produced by filtering an appropriate
volume of diluted cow faeces to provide 100 to 200 colonies following incubation. |
| Step 5. |
Colonies are replicated from the master grid onto grids
placed on chromogenic agar for presumptive identification of E. coli. |
| Step 6. |
Colonies are replicated from the master grid onto grids
placed on agar containing antimicrobials at National Centre for Clinical
Laboratory Standards (NCCLS) recommended concentrations. A copy is also
placed on agar containing no antimicrobials. |
| Step 7. |
Chromogenic, antimicrobial and control agar plates are
incubated overnight. |
| Step 8. |
Consistent interpretation of colony growth is achieved
by capturing digital images of HGMF grids and analysing using specific
software. |
| Step 9. |
Collation and standardised reporting of single and multiple
resistance traits is achieved within the software. |
Top of page
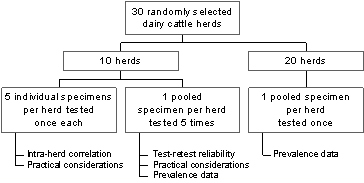
The advantages of the HGMF approach are that it can be used to appraise single and multiple resistance traits of up to 200 colonies per specimen. The use of image analysis provides a standard interpretation of results and avoids errors encountered with manual data recording. In the New South Wales study this has enabled deployment of a study design for deriving variance components followed by calculation of intracluster (intra-herd) correlation. The latter is useful for quantifying the extent of likeness within groups (in this case the clustering of E. coli resistance trait within herds of cattle) which impinges on the interpretation and analysis of data and is of interest in the design of future studies.10 The study will also provide prevalence estimates (proportion of isolates, proportion of herds) for single and multiple resistance traits for four antibiotics (gentamycin, ampicillin, tetracycline and sulfamethoxazole) at NCCLS 'intermediate' concentrations9 based on observations made on approximately 10,000 isolates from 30 randomly-selected dairy herds. Test-retest reliability11 is also being evaluated on pooled faecal samples from 20 dairy farms. Overall, the study will help generate the statistical assumptions required for a more comprehensive survey of livestock. The Figure summarises the design of this study.
Figure. Design of the pilot study for assessing prevalence of resistance, intra-herd clustering, and test-retest reliability
Top of page
Conventional approaches to surveillance for antimicrobial resistance in livestock and food usually rely on disk diffusion, agar dilution or broth dilution techniques. These methods are suited to comprehensive screening of individual isolates against a large number of drugs and drug concentrations. They are often chosen because they can provide data for commensals, animal pathogens, and zoonotic pathogens that can be compared to the data being obtained for human pathogens. Furthermore, because many technicians are familiar with these methods they are a convenient basis for standardisation of measurement systems. However, because these techniques are costly on a per isolate basis they are less appealing for ecological and population based studies that demand the evaluation of resistance traits of a large number of indicator organisms, such as E. coli.
Although there is no limit to the number of antibiotics or concentrations that may be evaluated in the HGMF resistance test, restricting the number of antibiotics avoids the difficulty of having to interpret data for a large number of resistance patterns (if a is the number of antibiotics evaluated then the test produces information on 2a resistance patterns). HGMF resistance testing is therefore suited to screening a very large number of isolates against a panel of the most important antibiotics. It is attractive to combine HGMF testing with other research on the biology of resistance by removing a sub-sample of screened isolates for more detailed analysis by conventional resistance tests or molecular techniques.
Top of page
References
1. Wray C, Gnanou JC. Antibiotic resistance monitoring in bacteria of animal origin: analysis of national monitoring programmes. Int J Antimicrob Agents 2000;14:291-294.
2. Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS system for mixed models. Cary, N.C.: SAS Institute Inc.; 1996.
3. Dunlop RH, McEwen SA, Meek AH, Clarke RC, Friendship RM, Black WD et al. Measurement of antimicrobial-resistant Escherichia coli in pig feces with a hydrophobic grid membrane filter interpreter system. Appl Environ Microbiol 1998;64:366-369.
4. Barton MD, Wilkins J. Antibiotic resistance in bacteria isolated from poultry. Canberra: Rural Industries Research and Development Corporation; 2001.
5. Barger F Editor. DANMAP 99 -Consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, food and humans in Denmark. Copenhagen: Danish Veterinary Laboratory; 1999.
6. Bengtsson B, Franklin A, Greko C, Karlsson MWC. SVARM 2000 -Swedish veterinary antimicrobial resistance monitoring. Uppsala: National Veterinary Institute, Sweden.
7. Bohning D, Greiner M. Prevalence estimation under heterogeneity in the example of bovine trypanosomosis in Uganda. Prev Vet Med 1998;36:11-23.
8. Dunlop RH, McEwen SA, Meek AH, Friendship RM, Black WD, Clarke RC. Sampling considerations for herd-level measurement of faecal Escherichia coli antimicrobial resistance in finisher pigs. Epidemiol Infect 1999;122:485-496.
9. National Centre for Clinical Laboratory Standards, Document M07-A5 'Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically', Approved Standard. 5th edition; 2000.
10. Baskerville NB, Hogg W, Lemelin J. The effect of cluster randomization on sample size in prevention research. J Fam Pract 2001;50:W241-W246.
11. Streiner DL, Norman GR. Health Measurement Scales. Second edition. Oxford, UK: Oxford University Press; 1995:231.
Top of page
Author affiliations
Correspondence: Dr David Jordan, Senior Research Scientist, New South Wales Agriculture, Bruxner Highway Wollongbar NSW 2477. Telephone: +61 2 6626 1240. Facsimile: +61 2 6628 1744. Email: david.jordan@agric.nsw.gov.au
This article was published in Communicable Diseases Intelligence Volume 27 Suppl, May 2003.
